Using machine learning to predict prostate cancer behavior
Prostate cancer is among the most common cancers in American men, accounting for one in five new cancer diagnoses. Hormone therapy is currently the standard of care for patients with metastatic disease, but nearly all patients develop resistance to this treatment eventually. Extensive effort has therefore been directed toward the search for new drug targets, illuminating the biological underpinnings of the disease. Given a tumor sample from a patient with prostate cancer, researchers can now identify millions of genetic and molecular features, from single DNA mutations to RNA transcription errors to mutant protein complexes.
Using this data to predict disease course, however, remains a challenge for clinicians. Such prognostic ability would require finding patterns in these millions of datapoints: which mutations impact which pathways, and which pathways lead to aggressive disease? There are many things humans do better than machines, but when a massive dataset beckons, computers—with the help of scientists—can learn fast.
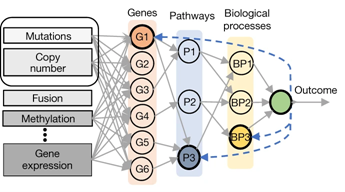
In this case, the scientists at the helm are former Clinical Investigator Eliezer M. Van Allen, MD, and his lab at Dana-Farber Cancer Institute. Last month, the team unveiled P-NET, a “biologically informed deep learning model” capable of guessing a prostate cancer patient’s disease state (primary or metastatic) given only the tumor’s mutated genes and how many copies of specific genes it had. Using pattern-recognition and error-correcting methods modeled after how the human brain learns, P-NET was able to predict more accurately and with far less information than any previous models. With further “learning,” P-NET could be used in the clinic to predict aggressive disease in patients with primary prostate cancer.
From P-NET’s output, the researchers were also able to form hypotheses about which genetic alterations and biological processes contribute to metastasis. The data implicated some already-known prostate cancer drivers, such as TP53, but also unexpected genes, such as MDM4, which was found to be overexpressed in the metastatic samples. By generating new knowledge about how prostate cancer progresses, the model may inform much-needed novel therapeutic strategies.
Dr. Van Allen’s work demonstrates the enormous potential at the intersection of cancer research and computational science. As genetic and molecular profiling becomes increasingly routine, P-NET shows us how machine learning can be leveraged to make sense of all that data—in prostate cancer and beyond.
Read more in Nature.
