A new way to visualize antibody-virus data
If you asked a hundred people to rate a hundred movies, you would generate enough data to be able to make some predictions. Someone who enjoyed Notting Hill would likely enjoy Pretty Woman, for instance; the new Guardians of the Galaxy movie will likely be a hit with longtime Marvel fans. This is an example of a bipartite dataset, which measures interactions between two types of entries—in this case, movies and viewers—and can be used not only to predict unmeasured interactions but also to reveal the underlying rules governing a system.
Because each measurement only conveys the relationship between two entries, transforming a set of these measurements into cohesive map of the whole system is a complex math problem, but it can be done. To use a more visual example, consider how the below table, which measures the similarity between two colors, is transformed into the canonical color wheel.
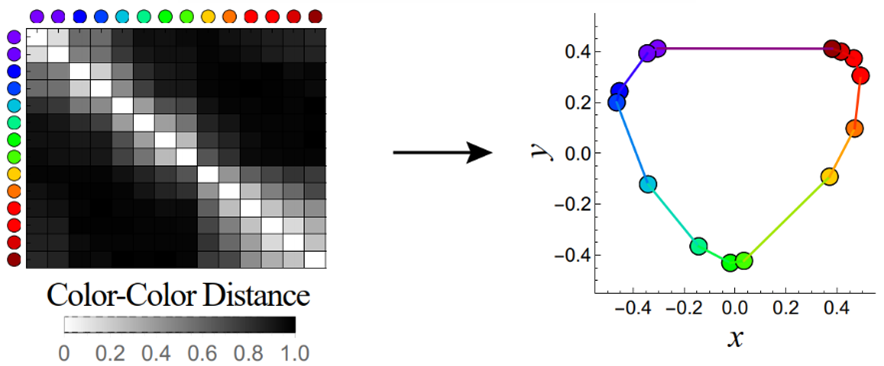
At Fred Hutchinson Cancer Center, Damon Runyon Quantitative Biology Fellow Tal Einav, PhD, and his colleagues are applying this methodology to antibody-virus datasets to uncover the rules governing these interactions. By mapping the distance between antibodies and viruses—where “distance” refers to how effectively an antibody neutralizes a virus—Dr. Einav and his team have gleaned valuable insight into antibody behavior.
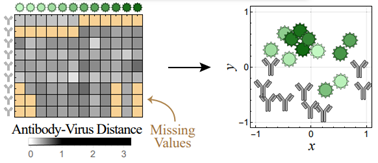
For example, they can predict how an existing antibody will respond to a new virus strain based on its previous interactions. They can also guide the design of optimized antibody cocktails. Notably, the team found that “most two-antibody cocktails can be mimicked by a single antibody, whereas cocktails with three or more antibodies often exhibit novel behavior that no single antibody can replicate.”
While Dr. Einav and his team used influenza virus data for this study, it is easy to see how their methods could be applied to predict a patient’s response to a new COVID-19 variant or to cancer-causing viruses like human papillomavirus. More broadly, their findings demonstrate how large datasets—increasingly the norm in biological studies—can be visualized to reveal underlying rules of the system and inform predictions about a disease agent’s response to treatment.
This research was published in Physical Review X.
