Genetic drivers of rare lymphoma identified, including two new cancer genes
A range of genetic disturbances can result in the same type of cancer, the way an off-tasting dish might result from any number of bad ingredients or missteps in the preparation process. Often, variation in clinical features—tumor appearance, location, behavior—is what defines cancer subtypes, while the genetic origins of each subtype remain unclear. But to make sense of this variation, and thus refine diagnosis and develop more precise treatments, researchers must trace these clinical features back to their genetic origins.
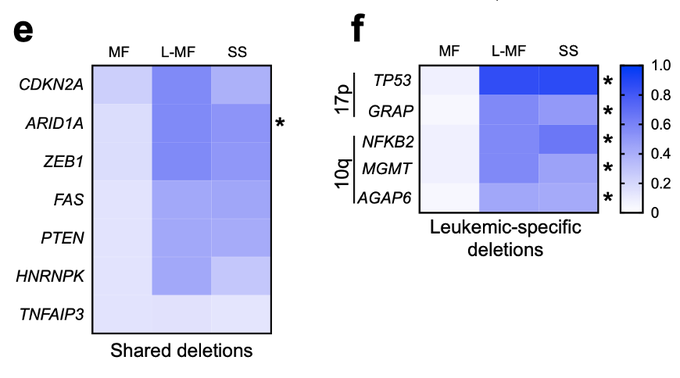
At Northwestern University, Damon Runyon Clinical Investigator Jaehyuk Choi, MD, PhD, and colleagues have launched such an investigation into cutaneous T cell lymphoma (CTCL), a rare form of non-Hodgkin lymphoma that arises when abnormalities in certain white blood cells cause them to attack the skin. The most common CTCL subtype, called mycosis fungoides (MF), presents as an itchy rash of varying size and severity. Patients with MF tend to have an excellent prognosis if treated early, but its resemblance to other skin conditions often impedes diagnosis. In rare cases the cancer will spread to the blood, and these patients are diagnosed with Sézary syndrome (SS). What determines a patient’s subtype, or how aggressive the disease will be, is not well understood on a molecular level.
(MF, leukemic-MF, and SS)
By sequencing thousands of CTCL patient samples across subtypes and stages, Dr. Choi’s team has illuminated numerous pathways from genetic mutation to clinical outcome, offering new potential targets for therapy as well as improved prognostic metrics. In total, the team uncovered 86 different “driver genes” that can be mutated in CTCL, including 19 genes not previously associated with this disease and two not previously associated with any cancer. Several of these mutations amplify the signal that tells immune T cells to multiply, underscoring a common pathway for lymphoma development that future therapies might interrupt.
While many driver genes were shared between MF and SS, the researchers found some that were subtype-unique. For example, the crucial tumor suppressor gene TP53 was deleted in 87% of SS samples, but in only 7% of MF samples. The team was also able to identify genetic predictors of disease severity. Most notably, patients who had a mutation in the PD1 protein, which sits on the surface of T cells and receives the “turn off” signal, faced a significantly worse prognosis.
With the genetics of CTCL newly clarified, Dr. Choi’s team has paved the way for screenings that predict disease course and therapies that target the disease’s many paths. They have also demonstrated the value of whole-genome sequencing in the study of cancers with wide clinical variation: sometimes there are not one, not two, but 86 missteps to avoid.
Learn more in Blood.
